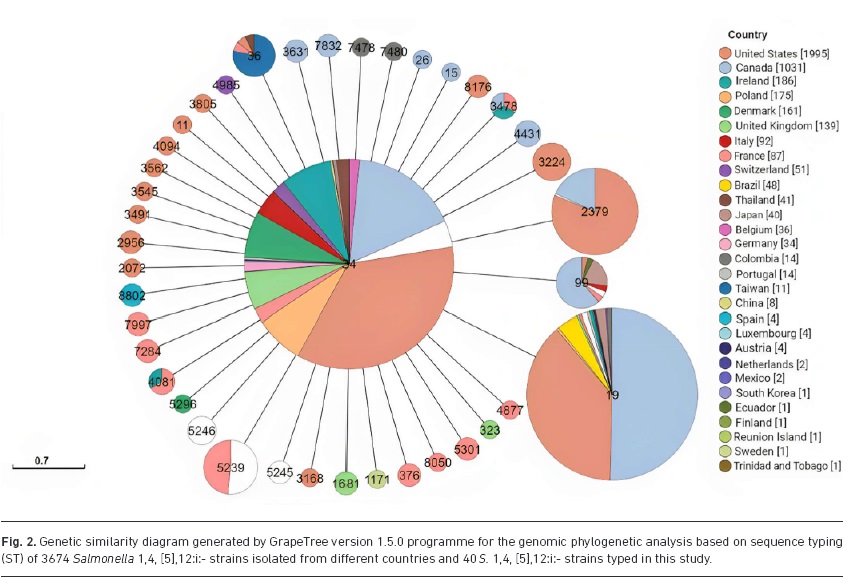
 Abstract: Introduction. Salmonella 1,4, [5],12:i:- strains with different antimicrobial resistance profiles have been associated with foodborne disease outbreaks in several countries. In Brazil, S. 1,4, [5],12:i:- was identified as one of the most prevalent serovars in São Paulo State during 2004-2020.Gap Statement. However, few studies have characterized this serovar in Brazil.Aim. This study aimed to determine the antimicrobial resistance profiles of S. 1,4, [5],12:i:- strains isolated from different sources in Southeast Brazil and compare their genetic diversity.Methodology. We analysed 113 S. 1,4, [5],12:i:- strains isolated from humans (n=99), animals (n=7), food (n=5) and the environment (n=2) between 1983 and 2020. Susceptibility testing against 13 antimicrobials was performed using the disc diffusion method for all the strains. Plasmid resistance genes and mutations in the quinolone resistance-determining regions were identified in phenotypically fluoroquinolone-resistant strains. Molecular typing was performed using enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) for all strains and multilocus sequence typing (MLST) for 40 selected strains. Results. Of the 113 strains, 54.87 % were resistant to at least one antimicrobial. The highest resistance rates were observed against ampicillin (51.33 %), nalidixic acid (39.82 %) and tetracycline (38.05 %). Additionally, 39 (34.51 %) strains were classified as multidrug-resistant (MDR). Nine fluoroquinolone-resistant strains exhibited the gyrA mutation (Ser96?Tyr96) and contained the qnrB gene. The 113 strains were grouped into two clusters using ERIC-PCR, and most of strains were present in one cluster, with a genetic similarity of =80 %. Finally, 40 strains were typed as ST19 using MLST.Conclusion. The prevalence of MDR strains is alarming because antimicrobial treatment against these strains may lead to therapeutic failure. Furthermore, the ERIC-PCR and MLST results suggested that most strains belonged to one main cluster. Thus, a prevalent subtype of Salmonella 1,4, [5],12:i:- strains has probably been circulating among different sources in São Paulo, Brazil, over decades. Abstract: Introduction. Salmonella 1,4, [5],12:i:- strains with different antimicrobial resistance profiles have been associated with foodborne disease outbreaks in several countries. In Brazil, S. 1,4, [5],12:i:- was identified as one of the most prevalent serovars in São Paulo State during 2004-2020.Gap Statement. However, few studies have characterized this serovar in Brazil.Aim. This study aimed to determine the antimicrobial resistance profiles of S. 1,4, [5],12:i:- strains isolated from different sources in Southeast Brazil and compare their genetic diversity.Methodology. We analysed 113 S. 1,4, [5],12:i:- strains isolated from humans (n=99), animals (n=7), food (n=5) and the environment (n=2) between 1983 and 2020. Susceptibility testing against 13 antimicrobials was performed using the disc diffusion method for all the strains. Plasmid resistance genes and mutations in the quinolone resistance-determining regions were identified in phenotypically fluoroquinolone-resistant strains. Molecular typing was performed using enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) for all strains and multilocus sequence typing (MLST) for 40 selected strains. Results. Of the 113 strains, 54.87 % were resistant to at least one antimicrobial. The highest resistance rates were observed against ampicillin (51.33 %), nalidixic acid (39.82 %) and tetracycline (38.05 %). Additionally, 39 (34.51 %) strains were classified as multidrug-resistant (MDR). Nine fluoroquinolone-resistant strains exhibited the gyrA mutation (Ser96?Tyr96) and contained the qnrB gene. The 113 strains were grouped into two clusters using ERIC-PCR, and most of strains were present in one cluster, with a genetic similarity of =80 %. Finally, 40 strains were typed as ST19 using MLST.Conclusion. The prevalence of MDR strains is alarming because antimicrobial treatment against these strains may lead to therapeutic failure. Furthermore, the ERIC-PCR and MLST results suggested that most strains belonged to one main cluster. Thus, a prevalent subtype of Salmonella 1,4, [5],12:i:- strains has probably been circulating among different sources in São Paulo, Brazil, over decades. |
| Journal of Medical Microbiology |
| v. 73, n. 2, p. 001792-1-001792-15 - Ano: 2024 |
| Fator de Impacto: 3,0 |
| |
 @article={003183327,author = {PEREIRA, Giovana do Nascimento; SERIBELLI, Amanda Aparecida; CAMPIONI, Fábio; GOMES, Carolina Nogueira; TIBA-CASAS, Monique Ribeiro; MEDEIROS, Marta Inês Cazentini; RODRIGUES, Dália dos Prazeres; FALCÃO, Juliana Pfrimer.},title={High levels of multidrug-resistant isolates of genetically similar Salmonella 1,4, [5],12:I: from Brazil between 1983 and 2020},journal={Journal of Medical Microbiology},note={v. 73, n. 2, p. 001792-1-001792-15},year={2024}} @article={003183327,author = {PEREIRA, Giovana do Nascimento; SERIBELLI, Amanda Aparecida; CAMPIONI, Fábio; GOMES, Carolina Nogueira; TIBA-CASAS, Monique Ribeiro; MEDEIROS, Marta Inês Cazentini; RODRIGUES, Dália dos Prazeres; FALCÃO, Juliana Pfrimer.},title={High levels of multidrug-resistant isolates of genetically similar Salmonella 1,4, [5],12:I: from Brazil between 1983 and 2020},journal={Journal of Medical Microbiology},note={v. 73, n. 2, p. 001792-1-001792-15},year={2024}} |





 Abstract: Introduction. Salmonella 1,4, [5],12:i:- strains with different antimicrobial resistance profiles have been associated with foodborne disease outbreaks in several countries. In Brazil, S. 1,4, [5],12:i:- was identified as one of the most prevalent serovars in São Paulo State during 2004-2020.Gap Statement. However, few studies have characterized this serovar in Brazil.Aim. This study aimed to determine the antimicrobial resistance profiles of S. 1,4, [5],12:i:- strains isolated from different sources in Southeast Brazil and compare their genetic diversity.Methodology. We analysed 113 S. 1,4, [5],12:i:- strains isolated from humans (n=99), animals (n=7), food (n=5) and the environment (n=2) between 1983 and 2020. Susceptibility testing against 13 antimicrobials was performed using the disc diffusion method for all the strains. Plasmid resistance genes and mutations in the quinolone resistance-determining regions were identified in phenotypically fluoroquinolone-resistant strains. Molecular typing was performed using enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) for all strains and multilocus sequence typing (MLST) for 40 selected strains. Results. Of the 113 strains, 54.87 % were resistant to at least one antimicrobial. The highest resistance rates were observed against ampicillin (51.33 %), nalidixic acid (39.82 %) and tetracycline (38.05 %). Additionally, 39 (34.51 %) strains were classified as multidrug-resistant (MDR). Nine fluoroquinolone-resistant strains exhibited the gyrA mutation (Ser96?Tyr96) and contained the qnrB gene. The 113 strains were grouped into two clusters using ERIC-PCR, and most of strains were present in one cluster, with a genetic similarity of =80 %. Finally, 40 strains were typed as ST19 using MLST.Conclusion. The prevalence of MDR strains is alarming because antimicrobial treatment against these strains may lead to therapeutic failure. Furthermore, the ERIC-PCR and MLST results suggested that most strains belonged to one main cluster. Thus, a prevalent subtype of Salmonella 1,4, [5],12:i:- strains has probably been circulating among different sources in São Paulo, Brazil, over decades.
Abstract: Introduction. Salmonella 1,4, [5],12:i:- strains with different antimicrobial resistance profiles have been associated with foodborne disease outbreaks in several countries. In Brazil, S. 1,4, [5],12:i:- was identified as one of the most prevalent serovars in São Paulo State during 2004-2020.Gap Statement. However, few studies have characterized this serovar in Brazil.Aim. This study aimed to determine the antimicrobial resistance profiles of S. 1,4, [5],12:i:- strains isolated from different sources in Southeast Brazil and compare their genetic diversity.Methodology. We analysed 113 S. 1,4, [5],12:i:- strains isolated from humans (n=99), animals (n=7), food (n=5) and the environment (n=2) between 1983 and 2020. Susceptibility testing against 13 antimicrobials was performed using the disc diffusion method for all the strains. Plasmid resistance genes and mutations in the quinolone resistance-determining regions were identified in phenotypically fluoroquinolone-resistant strains. Molecular typing was performed using enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) for all strains and multilocus sequence typing (MLST) for 40 selected strains. Results. Of the 113 strains, 54.87 % were resistant to at least one antimicrobial. The highest resistance rates were observed against ampicillin (51.33 %), nalidixic acid (39.82 %) and tetracycline (38.05 %). Additionally, 39 (34.51 %) strains were classified as multidrug-resistant (MDR). Nine fluoroquinolone-resistant strains exhibited the gyrA mutation (Ser96?Tyr96) and contained the qnrB gene. The 113 strains were grouped into two clusters using ERIC-PCR, and most of strains were present in one cluster, with a genetic similarity of =80 %. Finally, 40 strains were typed as ST19 using MLST.Conclusion. The prevalence of MDR strains is alarming because antimicrobial treatment against these strains may lead to therapeutic failure. Furthermore, the ERIC-PCR and MLST results suggested that most strains belonged to one main cluster. Thus, a prevalent subtype of Salmonella 1,4, [5],12:i:- strains has probably been circulating among different sources in São Paulo, Brazil, over decades. @article={003183327,author = {PEREIRA, Giovana do Nascimento; SERIBELLI, Amanda Aparecida; CAMPIONI, Fábio; GOMES, Carolina Nogueira; TIBA-CASAS, Monique Ribeiro; MEDEIROS, Marta Inês Cazentini; RODRIGUES, Dália dos Prazeres; FALCÃO, Juliana Pfrimer.},title={High levels of multidrug-resistant isolates of genetically similar Salmonella 1,4, [5],12:I: from Brazil between 1983 and 2020},journal={Journal of Medical Microbiology},note={v. 73, n. 2, p. 001792-1-001792-15},year={2024}}
@article={003183327,author = {PEREIRA, Giovana do Nascimento; SERIBELLI, Amanda Aparecida; CAMPIONI, Fábio; GOMES, Carolina Nogueira; TIBA-CASAS, Monique Ribeiro; MEDEIROS, Marta Inês Cazentini; RODRIGUES, Dália dos Prazeres; FALCÃO, Juliana Pfrimer.},title={High levels of multidrug-resistant isolates of genetically similar Salmonella 1,4, [5],12:I: from Brazil between 1983 and 2020},journal={Journal of Medical Microbiology},note={v. 73, n. 2, p. 001792-1-001792-15},year={2024}}